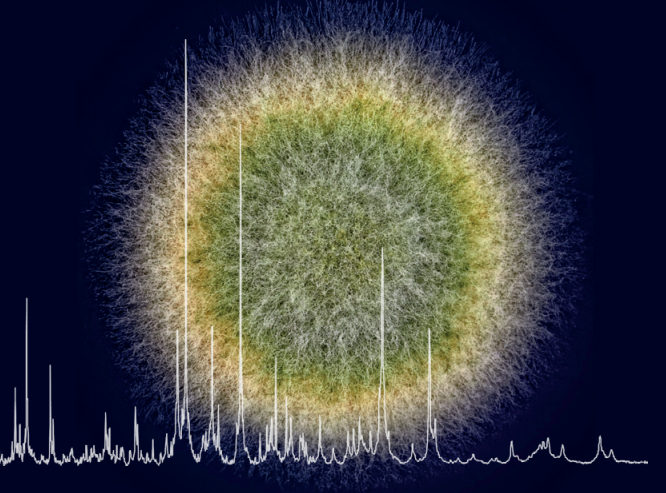
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-ToF MS) can be used to identify a number of fungi, down to species level. Sample testing involves growing cultures.
Moulds are more challenging to identify than yeasts, due to limitations in the number of spectra included in commercial libraries. For reviews see Wilkendorf et al (2020), Patel (2019) or Chalupová et al (2013).
Scroll down for video protocols, suppliers and test performance.
Video protocols

How it works
SAMPLE TYPES: Yeasts can be identified from culture plates, or directly from blood cultures. MALDI-ToF performs less well when blood samples contain mixed species of Candida or mixed bacterial/fungal cultures.
PERFORMING THE TEST: A small scraping from a positive culture is mixed with a specialised reagent matrix and a laser is used to ionise the sample, giving the particles electric charge. The masses of these ions are measured by the mass spectrometer. This takes approximately one minute and creates a spectrum. This is compared against an integrated database of reference strains and clinical isolates to provide identification. Calibrators and controls are regularly used to ensure high-quality performance of the MALDI-ToF mass spectrometer. For more detailed information about the mechanism visit the Shimadzu website.
LIMITATIONS: Filamentous fungi have usually been under-represented in commercial databases, whereas yeasts (especially Candida spp.) are well represented. This is beginning to change with new approaches to sampling increasing the efficacy of testing for filementous fungi (see Bruker’s Mycelium Transfer procedure). One of the challenges is that filamentous fungi often are quite variable in their growth pattern and therefore protein spectra may vary with media, time of growth and temperature.

Performance
ACCURACY: A systematic review by Xie et al (2019) reported an accuracy of 100% for most species of Candida (with the exception of C. dubliniensis and C. rugosa). For Aspergillus and other filamentous moulds (e.g. Mucor, Rhizopus, Lichtheimia), 82-97% of isolates were correctly identified to the species (complex) level.
TAT: Compared to standard methods, MALDI-ToF MS improves turn-around time for bacterial and yeast identification by an average of 1.5 days, and does not require subculture.
Melanised fungi (agents of phaeohyphomycosis) may give better results when grown in a liquid medium as this suppresses pigment formation. An optimised extraction protocol and expanded in-house database are described by Paul et al (2018).

Suppliers
SUPPLIERS: There are two commercial platforms currently approved in both Europe and the US: Bruker Biotyper (bacteria, yeast and filementous fungi) and BioMérieux VITEK MS (bacteria, yeast and filamentous fungi). The Axima@SARAMIS and Andromas are also approved in Europe.
Each comes with a proprietary database. The Bruker platform can be used in addition to other databases.
Conidia produces ID Fungi Plates (agar with a special membrane) that may help moulds and dermatophytes grow in a way that performs better in MALDI-ToF testing.

Libraries
Full species identification of fungi can be limited depending on the library that is used. In particular, filamentous fungi may be lacking in number of genera/species, or clinical strains. Dermatophytes are particularly difficult to identify.
Libraries may be tied to one particular platform, but most software allows the user to also build an in-house reference library from locally collected isolates.
The BCCM/IHEM database covers almost 250 genera of fungi and is publicly accessible via their web application.
Stein et al (2018) used the Bruker system to generate spectra that were analysed using 3 different libraries:
1) MSI 2016 library (Normand et al, 2017)
2) Bruker Filamentous Fungi Library v2.0.
3) NIH library database (Lau et al, 2013).
Using multiple libraries improved the detection rates, but around 25% of samples were not identified, even to genus level.